About BiSpark
BiSpark is a highly parallelized bisulfite-treated read aligner algorithm that utilizes distributed environment to significantly improve aligning performance and scalability. BiSpark is designed based on the Apache Spark distributed framework and shows highly efficient scalability.
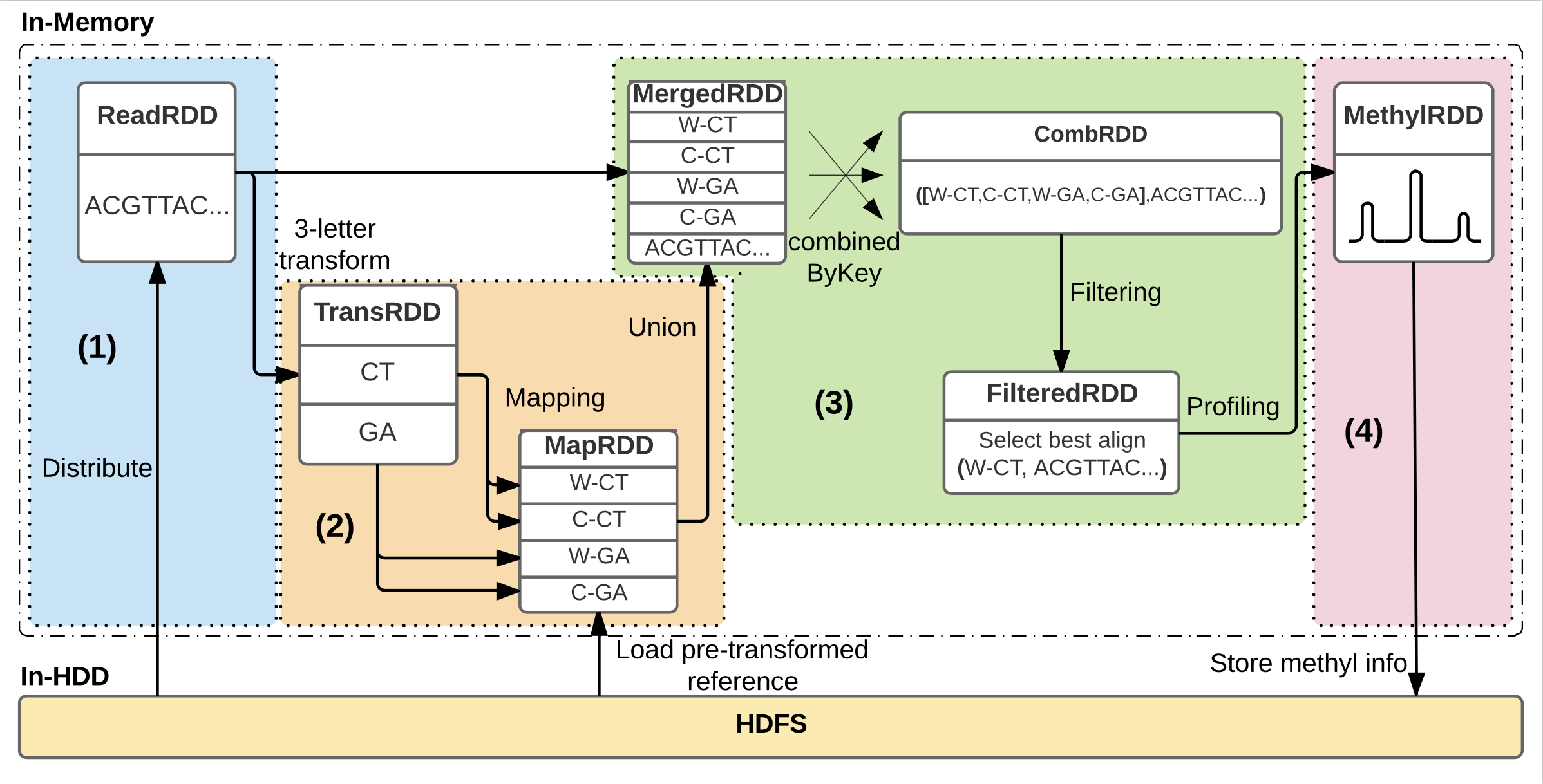
Analysis workflow within BiSpark consists of 4 processing phases: (1) Distributing the reads into key-value pairs, (2) Transforming reads into ‘three-letter’ reads and mapping to transformed reference genome, (3) Aggregating mapping results and filtering ambiguous reads, and (4) Profiling the methylation information for each read. The figure depicts the case when library of input data is a non-directional.
Features
- Fast and well scalable to both data size and cluster size.
- Based on Apache Spark: Easy to use. No restriction on the number of nodes.
- Using HDFS: Do not need to consider files to be distributed by user.
Installation
BiSpark is implemented on Apache Spark framework and HDFS file system. Bowtie2 is also used to alignment, thus all three frameworks and programs should be installed before running BiSpark. Bowtie2 should be callable on all slave nodes.
Requirements
- Python: 2.6, 2.7
- Apache Spark: >= 1.6
- HDFS: 2.6
Usage
- Building Index (reference: ./src/build_index/run.sh)
- –input: Input reference FASTA file located in HDFS.
- –output: Output directory for indexing files in HDFS.
- –log: Local log file path.
- Alignment (reference: ./src/alignment/run.sh)
- –input: Input reference FASTA file located in HDFS.
- –output: Output directory for indexing files in HDFS.
- –log: Local log file path.
- –ref: Index dicrectory in HDFS. Should be same path as denoted as ‘–output’.
- –nodes: The number of executors. = (# of nodes of cluster) * (# of spark executors in each node)
- –num_mm: The number of maximum mismatches (default: 4)
- –local_save: Local output sam file path.
- –testmode: Switching testmode for experiment. Should be one of ‘balancing’ or ‘plain’.
- –appname: Application name that are used for Spark’s application name.
Recommended (optional) pre-processing for quality control
To improve the mappability and alignment accuracy, snitizing the poor reads before the main BiSpark phase is highly recommended. Following is recommended procedure for read quality control.
- Run Cutadapt to remove adaptor sequence from the reads.
- Run FastQC to visualize the various aspects of your data including read quality.
Contact
If you have any question or problem, please send a email to dane2522@gmail.com