About DeepFam
DeepFam is a deep learning based alignment-free protein function prediction method. DeepFam first extracts features of conserved regions from a raw sequence by convolution layer and makes a prediction based on the features.
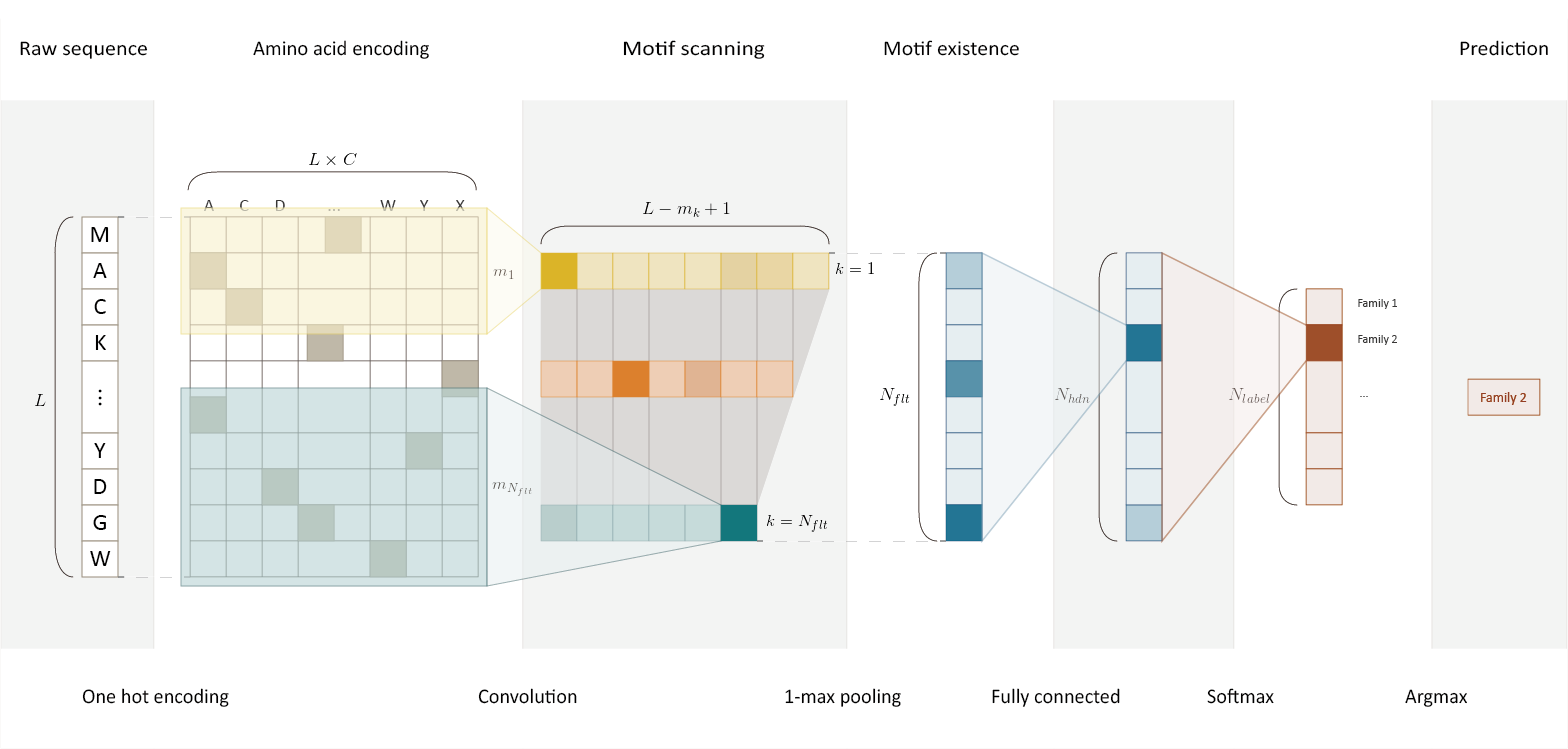
Features
- Alignment-free: Do not need multiple or pairwise sequence alignment to train family model.
- Instead, locally conserved regions within a family are trained by convolution units and 1-max pooling. Convolution unit works similar as PSSM.
- Utilizing variable-size convolution unit (multiscale convolution unit) to train family specific conserved regions whose lengths are usually various.
Installation
DeepFam is implemented in with Tensorflow library. Both CPU and GPU machines are supported. For detail instruction of installing Tensorflow, see the guide on official website.
Requirements
- Python: 2.7
- Tensorflow: over 1.0
Usage
First, clone the repository or download compressed source code files.
$ git clone https://github.com/bhi-kimlab/DeepFam.git
$ cd DeepFam
You can see the valid paramenters for DeepFam by help option:
$ python src/DeepFam/run.py --help
One example of parameter setting is like:
$ python src/DeepFam/run.py \
--num_windows [256, 256, 256, 256, 256, 256, 256, 256] \
--window_lengths [8, 12, 16, 20, 24, 28, 32, 36] \
--num_hidden 2000 \
--batch_size 100 \
--keep_prob 0.7 \
--learning_rate 0.001 \
--regularizer 0.001 \
--max_epoch 25 \
--seq_len 1000 \
--num_classes 1074 \
--log_interval 100 \
--save_interval 100 \
--log_dir '/tmp/logs' \
--test_file '/data/test.txt' \
--train_file '/data/train.txt'
Data
All data used by experiments described in manuscript is available at here.
Contact
If you have any question or problem, please send a email to dane2522@gmail.com